Markov chain Monte Carlo (MCMC) is a numerical method for approximating high-dimensional integrals and/or summations. The method was first described by (???) and later modified by (???). The algorithm that we will implement in this chapter is often referred to as the Metropolis-Hastings algorithm.
Use of the Metropolis-Hastings algorithm was mostly restricted to the field of statistical physics until it was discovered by statisticians in the mid 1980s (???). Statisticians, as a group, were largely unaware of the numerical method until (???) provided a description of the method as a way to approximate intractable probability densities. It is fair to say that MCMC has transformed the field of statistics, and in particular, Bayesian statistics.
Bayesian statisticians are interested in the posterior probability distribution of a parameter, \(\theta\), which can be calculated using Bayes’s theorem as \[ \Pr(\theta | D) = {\Pr(D | \theta) \, \Pr(\theta) \over \Pr(D)} \] Here, \(\Pr(\theta | D)\) is the posterior probability distribution of the parameter. The posterior probability is a conditional probability: the probability of the parameter conditioned on the observations, \(D\). The other factors in the equation include the likelihood [\(\Pr(D | \theta)\)], prior probability distribution of the parameter [\(\Pr(\theta)\)], and marginal likelihood [\(\Pr(D)\)].
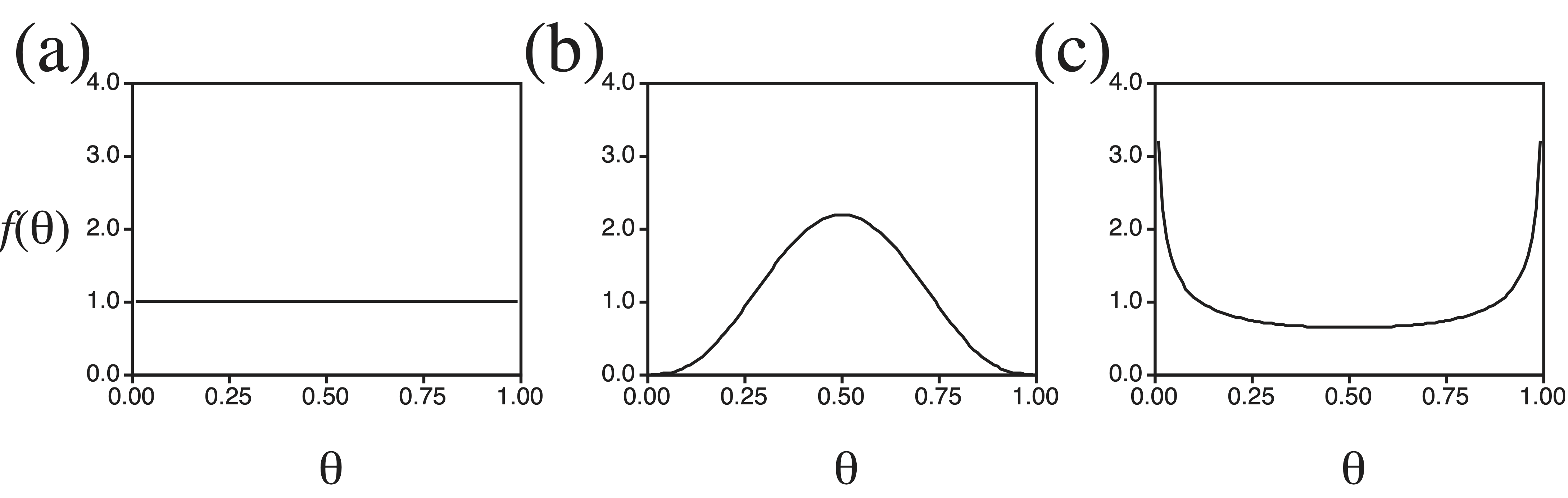
How does Bayesian analysis work in practice? Consider an experiment in which a coin is repeatedly tossed with the objective to estimate the probability that heads appears on a single toss of the coin, a parameter we call \(\theta\). We observe \(x\) heads on \(n\) tosses of the coin. In a Bayesian analysis, the objective is to calculate the posterior probability of the parameter, which for coin tossing is \[ f(\theta | x) = {f(x | \theta) f(\theta) \over \int_{0}^{1} f(x | \theta) f(\theta) \, d\theta} \] The likelihood, \(f(x | \theta)\), is given by the binomial probability distribution, \[ f(x | \theta) = {n \choose x} \theta^x (1-\theta)^{n-x} \] where the binomial coefficient is \({n \choose x} = {n! \over x! (n-x)!}\). In addition to the likelihood function, however, we must also specify a prior probability distribution for the parameter, \(f(\theta)\). This prior distribution should describe the investigator’s beliefs about the hypothesis before the experiment was performed. The problem, of course, is that different people may have different prior beliefs. In this case, it makes sense to use a prior distribution that is flexible, allowing different people to specify different prior probability distributions and also allowing for an easy investigation of the sensitivity of the results to the prior assumptions. A Beta distribution is often used as a prior probability distribution for the binomial parameter. The Beta distribution has two parameters, \(\alpha\) and \(\beta\). Depending upon the specific values chosen for \(\alpha\) and \(\beta\), one can generate a large number of different prior probability distributions for the parameter \(\theta\). Figure shows several possible prior distributions for coin tossing. Figure a shows a uniform prior distribution for the probability of heads appearing on a single toss of a coin. In effect, a person who adopts this prior distribution is claiming total ignorance of the dynamics of coin tossing. Figure b shows a prior distribution for a person who has some experience tossing coins; anyone who has tossed a coin realizes that it is impossible to predict which side will face up when tossed, but that heads appears about as frequently as tails, suggesting more prior weight on values around \(\theta = 0.5\) than on values near \(\theta = 0\) or \(\theta = 1\). Lastly, Figure c shows a prior distribution for a person who suspects he is being tricked. Perhaps the coin that is being tossed is from a friend with a long history of practical jokes, or perhaps this friend has tricked the investigator with a two-headed coin in the past. Figure c, then, might represent the ‘trick-coin’ prior distribution.

Besides being flexible, the Beta prior probability distribution has one other admirable property: when combined with a binomial likelihood, the posterior distribution also has a Beta probability distribution (but with the parameters changed). Prior distributions that have this property — that is, the posterior probability distribution has the same functional form as the prior distribution — are called conjugate priors in the Bayesian literature. The Bayesian treatment of the coin tossing experiment can be summarized as follows:
| Prior [\(f(\theta)\), Beta] | Likelihood [\(f(x | \theta)\), Binomial] | Posterior [\(f(\theta | x)\), Beta] |
|---|---|---|
| \({\Gamma(\alpha+\beta) \over \Gamma(\alpha) \Gamma(\beta)} \theta^{\alpha-1} (1-\theta)^{\beta-1}\) | \({n \choose x} \theta^x (1-\theta)^{n-x}\) | \({\Gamma(\alpha+\beta + n) \over \Gamma(\alpha + x) \Gamma(\beta + n - x)} \theta^{\alpha + x - 1} (1-\theta)^{\beta + n - x - 1}\) |
[The gamma function, not to be confused with the gamma probability distribution, is defined as \(\Gamma(y) = \int_{0}^{\infty} u^{y-1} e^{-u} \, du\). \(\Gamma(n) = (n - 1)!\) for integer \(n = 1, 2, 3, \ldots\) and \(\Gamma({1 \over 2}) = \pi\).] We started with a Beta prior distribution with parameters \(\alpha\) and \(\beta\). We used a binomial likelihood, and after some use of our undergraduate calculus we calculated the posterior probability distribution, which is also a Beta distribution but with parameters \(\alpha + x\) and \(\beta+n-x\).
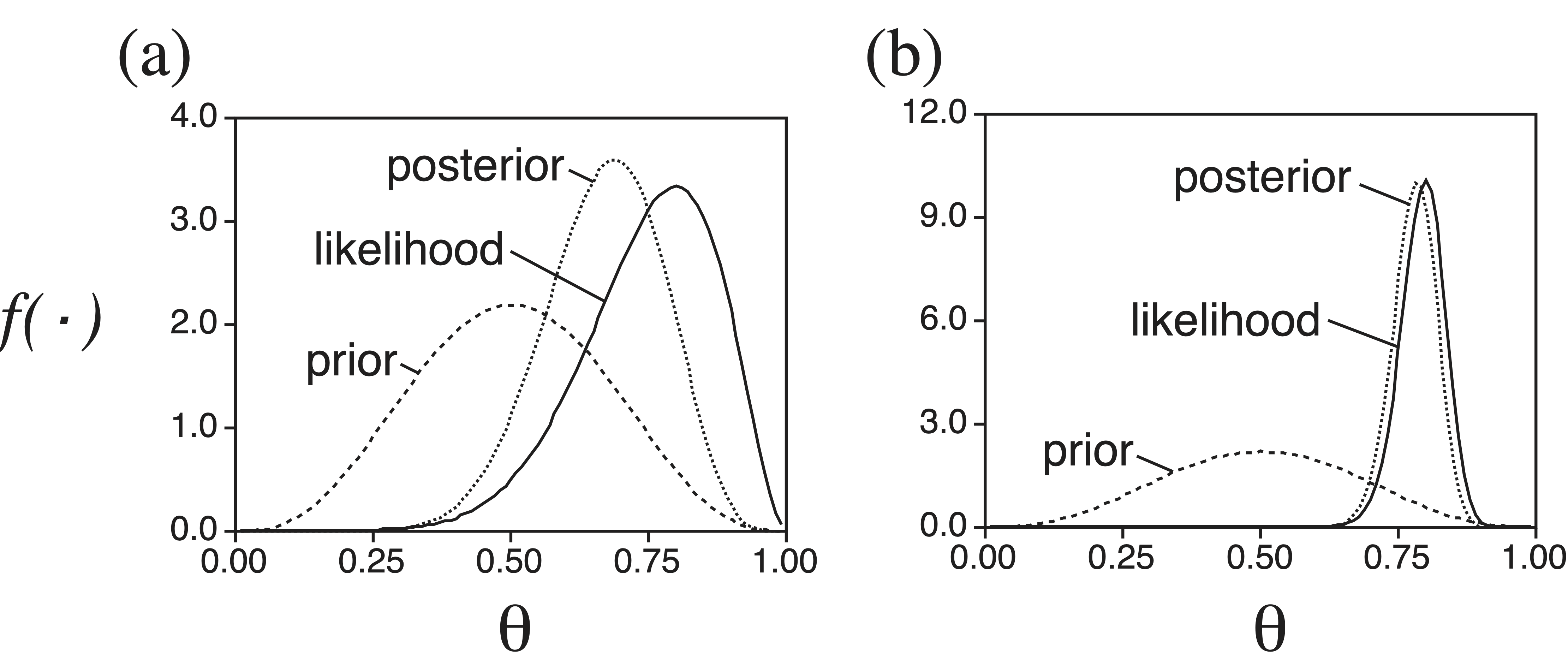
Figure 7.2 shows the relationship between the prior probability distribution, likelihood, and posterior probability distribution of \(\theta\) for two different cases, differing only in the number of coin tosses. The posterior probability of a parameter is a compromise between the prior probability and the likelihood. When the number of observations is small, as is the case for Figure 7.2a, the posterior probability distribution is similar to the prior distribution. However, when the number of observations is large, as is the case for Figure 7.2b, the posterior probability distribution is dominated by the likelihood.
Above, we casually claimed that the posterior distribution was a Beta distribution. Working out the math for this problem is informative. The posterior probability distribution for the coin tossing problem can be calculated analytically. Here, we assume a binomial probability distribution for the likelihood, and a Beta prior distribution on \(\theta\), which means the posterior probability distribution of the parameter \(\theta\) is: \[ f(\theta | x) = \frac{ {n \choose x} \theta^x (1-\theta)^{n-x} \frac{\Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta)} \theta^{\alpha-1} (1-\theta)^{\beta-1} }{ \int_{0}^{1} {n \choose x} \theta^x (1-\theta)^{n-x} \frac{\Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta)} \theta^{\alpha-1} (1-\theta)^{\beta-1} d\theta } \] Note that the denominator involves integration over all possible values of the parameter \(\theta\). Specifically, the probability of heads on a single toss of a coin, \(\theta\), can take values between 0 and 1.
Evaluating the integral in the denominator depends upon the following observation: The Beta probability distribution, like all continuous probability distributions, evaluates to one when integrated over all possible values for the parameter: \[\begin{eqnarray*} \int_{0}^{1} \frac{\Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta)} x^{\alpha-1} (1-x)^{\beta-1}dx & = & 1 \\ {\Gamma(\alpha+\beta) \over \Gamma(\alpha)\Gamma(\beta)} \int_{0}^{1} x^{\alpha-1} (1-x)^{\beta-1} dx & = & 1 \\ \int_{0}^{1} x^{\alpha-1} (1-x)^{\beta-1} dx & = & {\Gamma(\alpha)\Gamma(\beta) \over \Gamma(\alpha+\beta) } \end{eqnarray*}\] The denominator in the equation for the posterior probability distribution of \(\theta\), above, is \[\begin{eqnarray*} \int_{0}^{1} {n \choose x} \theta^x (1-\theta)^{n-x} {\Gamma(\alpha+\beta) \over \Gamma(\alpha)\Gamma(\beta)} \theta^{\alpha-1} (1-\theta)^{\beta-1} d\theta \\ \frac{ {n \choose x} \Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta) } \int_{0}^{1} \theta^x (1-\theta)^{n-x} \theta^{\alpha-1} (1-\theta)^{\beta-1} d\theta \\ \frac{ {n \choose x} \Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta) } \int_{0}^{1} \theta^{\alpha+x-1} (1-\theta)^{\beta+n-x-1} d\theta \end{eqnarray*}\] Because ${0}^{1} x^{-1} (1-x)^{-1} dx = ()() / (+) $, this means that ${0}^{1} x^{+x-1} (1-x)^{+n-x-1} dx = (+x)(+n-x) /(++ n) $. The posterior probability distribution for \(\theta\), then, is \[\begin{eqnarray*} f(\theta | x) & = & \frac{ {n \choose x} \theta^x (1-\theta)^{n-x} {\Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta)} \theta^{\alpha-1} (1-\theta)^{\beta-1} \over \frac{ {n \choose x} \Gamma(\alpha+\beta)}{\Gamma(\alpha)\Gamma(\beta) } { \Gamma(\alpha+x)\Gamma(\beta+n-x) \over \Gamma(\alpha+\beta + n) } }\\ & = & {\Gamma(\alpha+\beta+n) \theta^x (1-\theta)^{n-x} \theta^{\alpha-1} (1-\theta)^{\beta-1} \over \Gamma(\alpha+x)\Gamma(\beta+n-x)}\\ & = & { \Gamma(\alpha+\beta+n) \theta^{\alpha+x-1} (1-\theta)^{\beta+n-x-1} \over \Gamma(\alpha+x)\Gamma(\beta+n-x)} \end{eqnarray*}\] which is a Beta probability distribution with parameters \(\alpha+x\) and \(\beta+n-x\).
I will make one comment about the analytical solution of the posterior probability distribution for the coin-tossing problem: it involved hard math! Certainly, working through the math was at the edge of my ability. However, for problems that are only slightly more complicated than coin tossing (i.e., almost all other statistical problems), it is not possible to obtain the posterior probability distribution analytically. Until the advent of computers and algorithms for approximating high-dimensional integrals/summation, people were up a river without a paddle if their problem was somewhat complicated (i.e., problems that are actually interesting).
Even though we know the posterior probability distribution for the coin-tossing problem, we will act as if we don’t and approximate the posterior probability distribution using MCMC. The goal of MCMC is to construct a Markov chain that has as its state space the parameters of the model and a stationary distribution that is the posterior probability of the parameters. How do we construct a Markov chain that has these properties? The answer: Use the Metropolis-Hastings algorithm. The steps of the Metropolis-Hastings algorithm are as follows:
The steps of the Metropolis-Hastings algorithm are repeated many thousands, or millions, of times. The states that are visited form a Markov chain. The key to MCMC is that the sampled states are valid, albeit dependent, draws from the posterior probability distribution.
Create a new file called Model.h Work through the code below which needs to be added to the to file.
#ifndef Model_h
#define Model_h
#include <stdio.h>
#include <vector>
// forward declarations
class Alignment;
class ExponentialDistribution;
class PhyloCTMC;
class PureBirthProcess;
class RateMatrix_JC;
class Tree;
class UniformDistribution;
/**
* \class Model
*
* \brief This class represents the phylogenetic model.
*
*
*
* \author Sebastian Höhna
*
*/
class Model {
public:
Model(const std::string &fn);
virtual ~Model();
// parameters
double getBirthRate(void) const;
double getClockRate(void) const;
void setBirthRate( double r );
void setClockRate( double r );
// distributions
double getBirthRatePrior(void);
double getClockRatePrior(void);
double getLikelihood(void);
double getTreePrior(void);
private:
Alignment* my_alignment;
double* my_birth_rate;
double* my_clock_rate;
RateMatrix_JC* my_rate_matrix;
Tree* my_tree;
PureBirthProcess* tree_prior;
ExponentialDistribution* birth_rate_prior;
UniformDistribution* clock_rate_prior;
PhyloCTMC* likelihood;
};
#endif /* Model_h */Create a new file called Model.cpp Work through the code below which needs to be added to the to file.
#include "Model.h"
#include "Alignment.h"
#include "AlignmentReader.h"
#include "ExponentialDistribution.h"
#include "NewickTreeReader.h"
#include "PhyloCTMC.h"
#include "PureBirthProcess.h"
#include "RateMatrix_JC.h"
#include "Tree.h"
#include "UniformDistribution.h"
Model::Model(const std::string &fn)
{
// read in some trees
NewickTreeReader tree_reader = NewickTreeReader();
std::vector<Tree> trees = tree_reader.readTrees("primates.tre");
Tree* my_tree = new Tree(trees[0]);
// read in the data (i.e., the alignment)
AlignmentReader reader;
Alignment* my_alignment = new Alignment( reader.readPhylip( fn ) );
// create a transition rate matrix
my_rate_matrix = new RateMatrix_JC();
// create the clock rate prior
double* min_clock_rate = new double(0.0);
double* max_clock_rate = new double(100.0);
clock_rate_prior = new UniformDistribution(min_clock_rate, max_clock_rate);
my_clock_rate = new double(1.0);
clock_rate_prior->setValue( my_clock_rate );
// now set up the tree prior
double* birth_rate_mean = new double(1.0);
birth_rate_prior = new ExponentialDistribution(birth_rate_mean);
// specify the birth rate
my_birth_rate = new double(1.0);
birth_rate_prior->setValue( my_birth_rate );
tree_prior = new PureBirthProcess( my_alignment->getTaxonNames(), my_birth_rate );
tree_prior->setValue( my_tree );
likelihood = new PhyloCTMC( my_tree, my_rate_matrix, my_clock_rate );
likelihood->setValue( my_alignment );
}
Model::~Model()
{
delete my_alignment;
delete my_birth_rate;
delete my_clock_rate;
delete my_rate_matrix;
delete my_tree;
delete tree_prior;
delete birth_rate_prior;
delete clock_rate_prior;
delete likelihood;
}
double Model::getBirthRate(void) const
{
return *my_birth_rate;
}
double Model::getBirthRatePrior(void)
{
return birth_rate_prior->lnProbability();
}
double Model::getClockRate(void) const
{
return *my_clock_rate;
}
double Model::getClockRatePrior(void)
{
return clock_rate_prior->lnProbability();
}
double Model::getLikelihood(void)
{
return likelihood->lnProbability();
}
double Model::getTreePrior(void)
{
return tree_prior->lnProbability();
}
void Model::setBirthRate( double r )
{
*my_birth_rate = r;
}
void Model::setClockRate( double r )
{
*my_clock_rate = r;
}Create a new file called MCMC.h Work through the code below which needs to be added to the to file.
#ifndef MCMC_h
#define MCMC_h
#include <stdio.h>
#include <vector>
#include <string>
#include <iostream>
#include <fstream>
#include "Model.h"
#include "RandomNumberGenerator.h"
// forward declarations
/**
* \class MCMC
*
* \brief This class represents the MCMC algorithm.
*
*
*
* \author Sebastian Höhna
*
*/
class MCMC {
public:
MCMC(const std::string& filename);
MCMC(const MCMC& m);
virtual ~MCMC();
void run(int n_gen);
private:
// our moves
void monitorParameters(int g);
void updateBirthRate(void);
void updateClockRate(void);
Model my_model;
RandomNumberGenerator rng;
std::ofstream parameter_file;
std::ofstream tree_file;
};
#endif /* MCMC_h */Create a new file called MCMC.cpp Work through the code below which needs to be added to the to file.
#include "MCMC.h"
#include <math.h> /* isnan, sqrt */
MCMC::MCMC( const std::string &filename ) :
my_model( filename ),
parameter_file( "my_analysis.log" ),
tree_file( "my_analysis.trees" )
{
}
MCMC::MCMC( const MCMC &m ) :
my_model( "" ),
parameter_file( "my_analysis.log" ),
tree_file( "my_analysis.trees" )
{
}
MCMC::~MCMC( void )
{
// nothing to do here
}
void MCMC::monitorParameters( int gen )
{
parameter_file << gen;
parameter_file << "\t";
parameter_file << my_model.getBirthRate();
parameter_file << "\t";
parameter_file << my_model.getClockRate();
parameter_file << "\t";
parameter_file << std::endl;
}
void MCMC::updateBirthRate( void )
{
// get the current birth rate
double current_rate = my_model.getBirthRate();
// draw a new random uniform number
double u = rng.uniform01();
// define the window size
double delta = 7.32;
// now we compute the new proposed values
double new_rate = current_rate + (u-0.5) * delta;
// compute the prior ratio and likelihood ratio
double current_prior = my_model.getBirthRatePrior();
double current_likelihood = my_model.getTreePrior();
// set the new value
my_model.setBirthRate( new_rate );
double new_prior = my_model.getBirthRatePrior();
double new_likelihood = my_model.getTreePrior();
double acceptance_prob = new_prior - current_prior + new_likelihood - current_likelihood;
u = rng.uniform01();
if ( isnan(acceptance_prob) || u > acceptance_prob )
{
// reject -> reset the value
my_model.setBirthRate( current_rate );
}
else
{
// accept! nothing to do, because we changed our value already
}
}
void MCMC::updateClockRate( void )
{
}
void MCMC::run( int num_generations )
{
// we assume that the model and all parameters are initialized.
int thinning = 10;
for ( int current_gen=0; current_gen<num_generations; current_gen++ )
{
// draw new values:
updateBirthRate();
updateClockRate();
// now we monitor the variables
if ( current_gen % thinning == 0 )
{
monitorParameters( current_gen );
}
}
std::cout << "Hooray, we finished our MCMC run!" << std::endl;
}